About
I am a CS PhD student at Stanford University specializing in Convex Optimization and Probability. I am excited about applications of convex optimization to probabilistic challenges. I've previously worked on CS Theory, health, bio, and machine learning applications. In particular, we develop theoretical guarantees on temporal dependent data and software tools to work with Wearable devices website.
Publications
TD(0) Learning converges for Polynomial mixing and non-linear functions
A. Sridhar, A. Johansen
ArXiv, 2025
PDF
Abstract
Bibtex
SignalP 6.0 predicts all five types of signal peptides using protein language models
F. Teufel, J.J.A. Armenteros, A.R. Johansen, M.H. Gislason, S.I. Pihl, K.D. Tsirigos, O. Winther, S. Brunak, G.V. Heijne, H. Nielsen
Nature Biotechnology, 2022
PDF
Abstract
Bibtex
Website

Deep protein representations enable recombinant protein expression prediction
H.M. Martiny, J.J.A. Armenteros, A.R. Johansen, J. Salomon, H. Nielsen
Computational Biology and Chemistry, 2021
PDF
Abstract
Bibtex
Prediction of GPI-Anchored proteins with pointer neural networks
M.H. Gislason, H. Nielsen, J.J.A. Armenteros*, A.R. Johansen* (*equal contribution)
Current Research in Biotechnology, 2021
PDF
Abstract
Bibtex
Website
Short Term Blood Glucose Prediction based on Continuous Glucose Monitoring Data
A. Mohebbi, A.R. Johansen, N. Hansen, P.E. Christensen, J.M. Tarp, M.L. Jensen, H. Bengtsson, M. Mørup
IEEE EMBC, 2020
PDF
Abstract
Bibtex
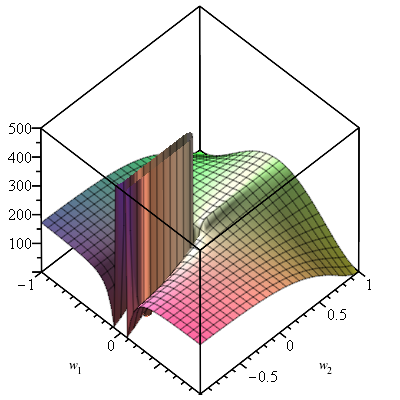
Neural Arithmetic Units
A. Madsen, A.R. Johansen
ICLR (Spotlight), 2020
PDF
Abstract
Bibtex
Website
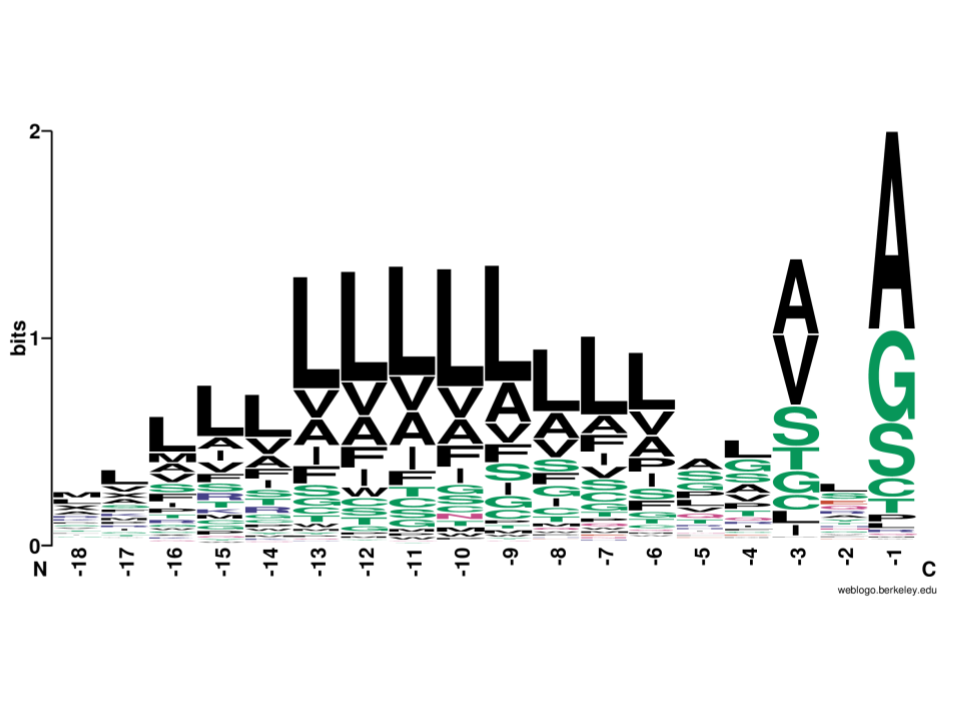
Language modelling for biological sequences curated datasets and baselines
J.J.A. Armenteros*, A.R. Johansen*, O. Winther, H. Nielsen (*equal contribution)
Preprint and website, 2019
PDF
Abstract
Bibtex
Website
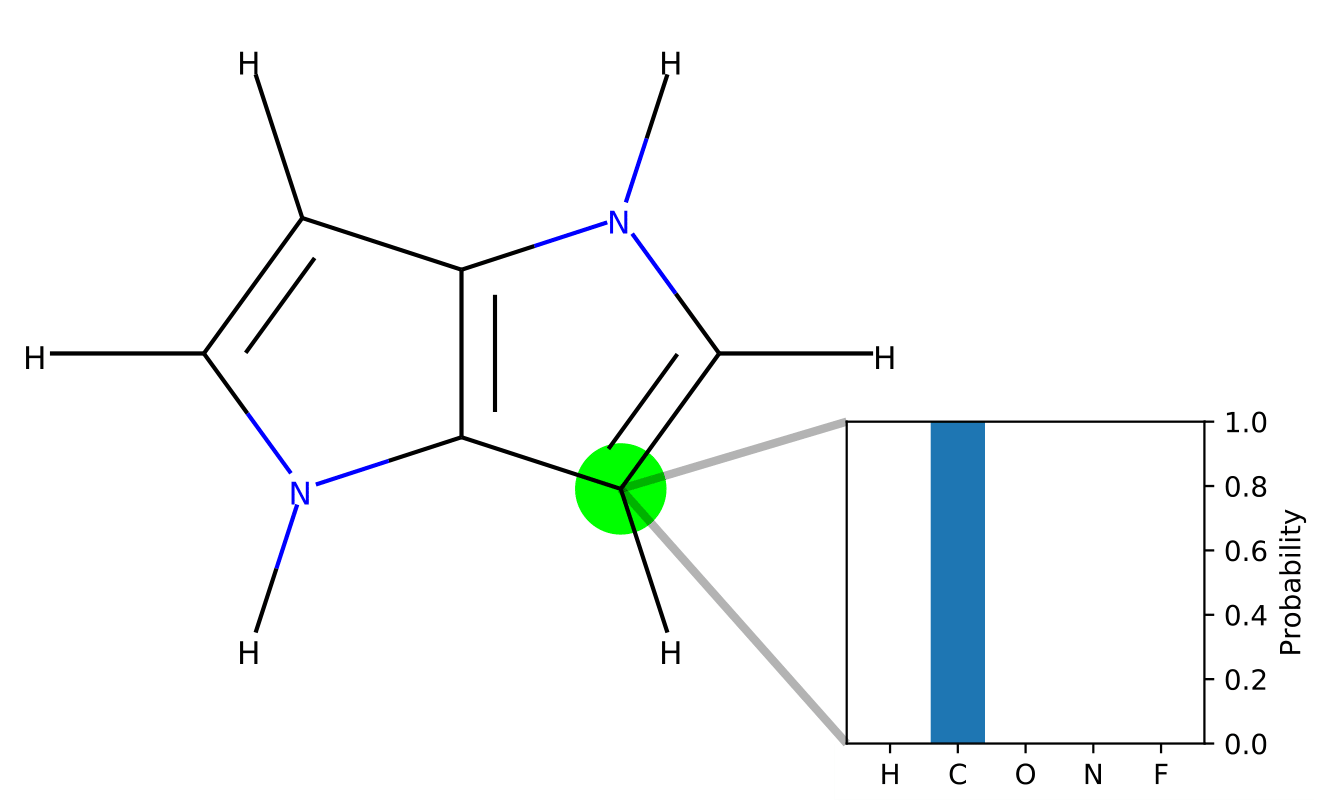
Autoencoding undirected molecular graphs with neural networks
J.J.W. Olsen, P.E. Christensen, M.H. Hansen, A.R. Johansen
Under review Journal of Chemical Information and Modeling, 2019
PDF
Abstract
Bibtex
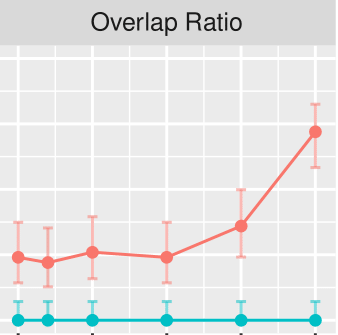
Measuring Arithmetic Extrapolation Performance
A. Madsen, A.R. Johansen
SEDL Workshop @ NeurIPS, 2019
PDF
Abstract
Bibtex
Code
Website
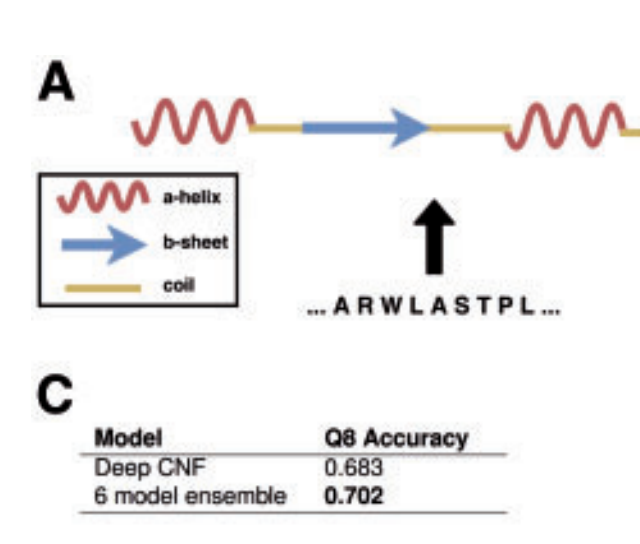
An introduction to deep learning on biological sequence data - examples and solutions
V.I. Jurtz, A.R. Johansen, M. Nielsen, J.J.A. Armenteros, H. Nielsen, C.K. Sønderby, O. Winther, S.K. Sønderby
Oxford Bioinformatics, 2017
Abstract
Bibtex
Code
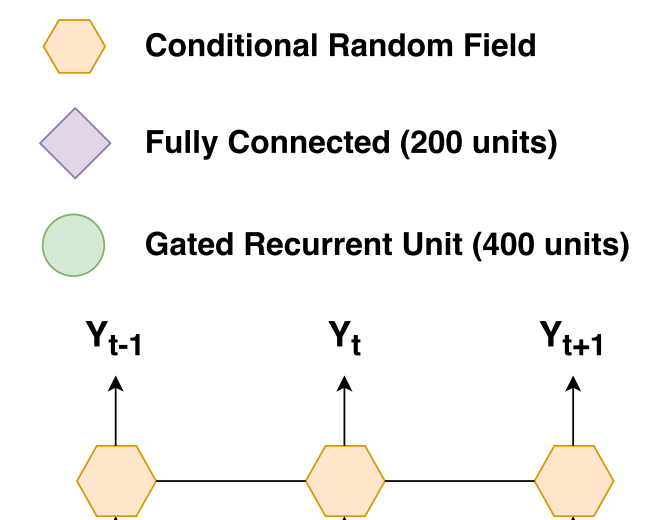
Deep Recurrent Conditional Random Field Network for Protein Secondary Prediction
A.R. Johansen, C.K. Sønderby, S.K. Sønderby, O. Winther
ACM BCB, 2017
PDF
Abstract
Bibtex
Code
A deep learning approach to adherence detection for type 2 diabetics
A. Mohebbi, T.B. Aradóttir, A.R. Johansen, H. Bengtsson, M. Fraccaro, and M. Mørup
IEEE EMBC, 2017
Abstract
Bibtex
Learning when to skim and when to read
A.R. Johansen, R. Socher
REPL4NLP Workshop @ ACL, 2017
PDF
Abstract
Bibtex
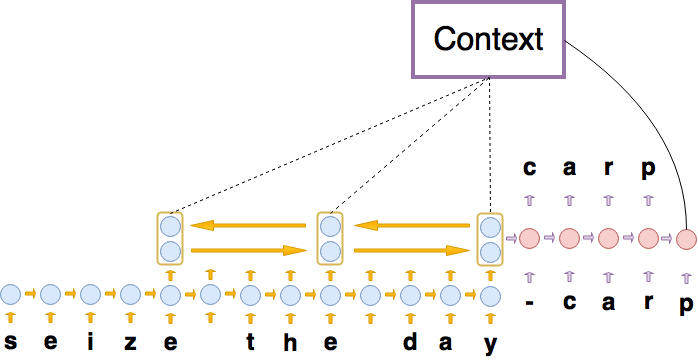
Neural Machine Translation with Characters and Hierarchical Encoding
A.R. Johansen, J.M. Hansen, E.K. Obeid, C.K. Sønderby, O. Winther
RNN Symposium @ NIPS, 2016
PDF
Abstract
Bibtex
Code
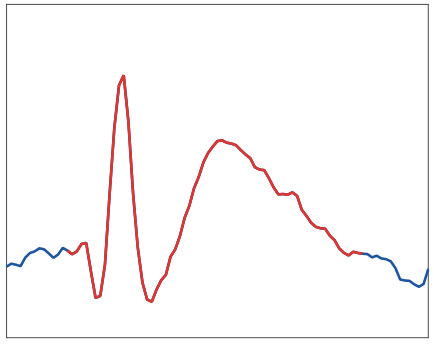
Epileptiform spike detection via convolutional neural networks
A.R. Johansen, J. Jin, T. Maszczyk, J. Dauwels, S.S. Cash, M.B. Westover
IEEE ICASSP, 2016
Abstract
Bibtex
Code
Supervising
At my time at the Technical University of Denmark together with Jose J.A. Armenteros I have supervised graduate students and independent researchers. This has continued after I have transitioned to my PhD at Stanford. If you are interested in joining our lab please write me an email with you resume and detail your background in machine learning. Below are listed titles of the M.Sc. Thesis' and independent research projects I have co-supervised.
Wearipedia
Includes approx 15 Stanford Undergraduate students.
2021-24
Predicting the thermostability of enzymes from protein sequences using neural networks and transfer learning
Gustav Lindved
2020
SignalP 6.0 achieves complete signal peptide prediction using deep protein representations
Felix Teufel
2020
Exploratory methods for annotating data structures by decompositions of difficult questions
Gabriel Enemark-Broholm and Marcus Skov Hansen
2020
Generalized autoregressive pretraining for improved understanding of proteins
Magnus Nolsøe and Frederik Wollesen Andersen
2019
Partially autoregressive language modelling and editing of discrete sequences' thesis
Daniel Horvath
2019
Prediction of sorting signals in eukaryotic proteins using deep learning
Magnús Halldór Gíslason
2019
Visual question answering using structured exploration and reinforcement learning
Jacob Johansen
2019
Evaluation of contextual molecular representations on predicting structural properties in binarized molecules
Jeppe Waarkjær Olsen
2019
Evaluation of contextual amino acid representations on the prediction of N-terminal targeting peptides based on deep learning
Silas Pihl
2019
Predicting Recombinant Gene Expression in Bacillus using Deep Learning Techniques
Hannah-Marie Martiny
2019
Evaluation of Pre-trained Amino Acid
Embeddings in Protein Prediction Tasks
Mikkel Møller Brusen and Gustav Madslund
2019
Code
Community
Previously, to build a strong deep learning community in Denmark I organized events for students with machine learning researchers and interested companies. This has led to 7 events with +1.5k student participants and sponsors such as Nordea, KPMG, Novozymes, Oticon, and Novo Nordisk. Post COVID I hope to make similar efforts in the bay area.
Deep Learning Copenhagen
Organizing events centered around maching learning technologies
2018-2020
Meetup
Teaching
Below you can find a list over courses I have taught in.
DTU course 02456 Deep learning
Head teaching assistant, designed programming exercises in PyTorch
Fall, 2019
Code
Website

DTU special course Deep reinforcement learning
Co-intructor, designed and evaluated course
June, 2019

DTU special course Introduction to reinforcement learning
Intructor, designed and evaluated course
Spring, 2019

DTU course 02456 Deep learning
Head teaching assistant, developed programming exercises in PyTorch
Fall, 2018
Code
Website

DTU course 02456 Deep learning
Teaching assistant, developed programming exercises in TensorFlow
Fall, 2016
Code
Website

Nvidia Deep Learning using TensorFlow
Exercise instructor, developed programming exercises in TensorFlow
September, 2016
Code